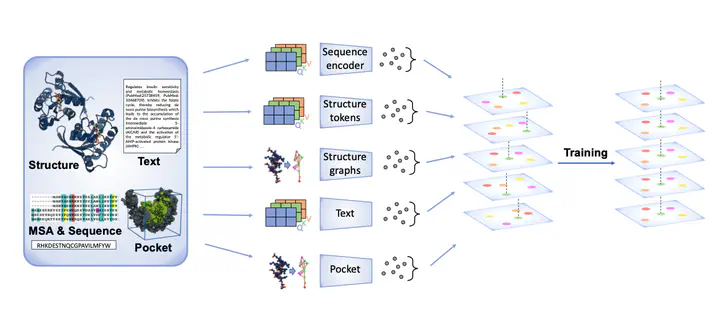
 Overview of the OneProt model. The model aligns multiple modalities, including primary protein sequence, 3D protein structure, binding pockets and text annotations. Each modality is processed by its respective encoder, generating embeddings that are then aligned in a shared latent space, facilitating cross-modal learning and integration.
Overview of the OneProt model. The model aligns multiple modalities, including primary protein sequence, 3D protein structure, binding pockets and text annotations. Each modality is processed by its respective encoder, generating embeddings that are then aligned in a shared latent space, facilitating cross-modal learning and integration.Abstract
Recent AI advances have enabled multi-modal systems to model and translate diverse information spaces. Extending beyond text and vision, we introduce OneProt, a multi-modal AI for proteins that integrates structural, sequence, alignment, and binding site data. Using the ImageBind framework, OneProt aligns the latent spaces of modality encoders along protein sequences. It demonstrates strong performance in retrieval tasks and surpasses state-of-the-art methods in various downstream tasks, including metal ion binding classification, gene-ontology annotation, and enzyme function prediction. This work expands multi-modal capabilities in protein models, paving the way for applications in drug discovery, biocatalytic reaction planning, and protein engineering.
The OneProt project demonstrates how multi-modal protein data—spanning sequences, structures, binding sites, and text annotations—can be integrated into a cohesive latent space using the ImageBind framework. This preprint details OneProt’s architecture, its alignment strategy, and its performance across tasks like enzyme function prediction and gene ontology annotation.
View the paper on arXiv or explore the OneProt code repository for implementation details.